Example: a single reaction¶
Consider the reaction 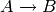 , where initially there
are 20 copies of the species
, where initially there
are 20 copies of the species 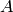 , and 0 copies of the
species
, and 0 copies of the
species 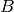 .
.
We will create a short CmePy script to solve the Chemical Master Equation for this reaction. Our script shall produce the following plot, which illustrates how the probability distribution over the reaction counts evolves in time:
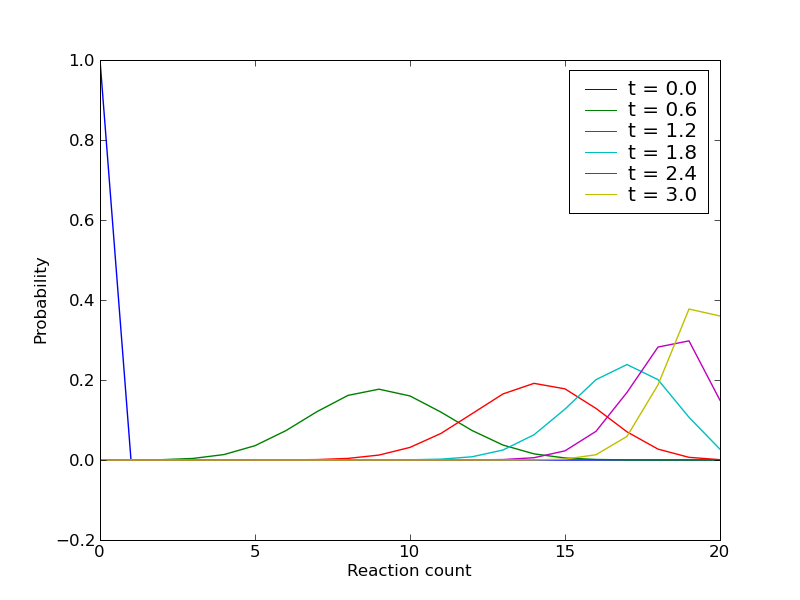
Observe how the probability begins concentrated at reaction count zero for
the time 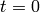 , quickly diffuses, then begins to accumulate at
reaction count 20, where all the copies of the species
, quickly diffuses, then begins to accumulate at
reaction count 20, where all the copies of the species 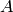 have been
exhausted.
have been
exhausted.
Defining the model¶
A CmePy script typically contains three main components
- A model, which defines the system of reactions.
- A solver, which solves the Chemical Master Equation (CME) for the model.
- A recorder, which stores solutions computed by the solver, and provides a simple interface for computing and plotting results, such as the expected value, standard deviation, or marginal distribution for species counts.
We begin by defining a model m for this simple system of reactions:
from cmepy import model
initial_copies = 20
m = model.create(
propensities = [lambda x: initial_copies - x],
transitions = [(1, )],
shape = (initial_copies + 1, ),
initial_state = (0, )
)
The reactions are specified by the propensities and transitions keyword
arguments. Since there is only a single reaction 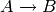 in this example, only one propensity function and transition are required.
The propensity function lambda x: initial_copies - x returns the propensity
of the reaction in terms of the reaction count x,
while the transition (1, ) specifies that the reaction increases the
reaction count x by 1.
in this example, only one propensity function and transition are required.
The propensity function lambda x: initial_copies - x returns the propensity
of the reaction in terms of the reaction count x,
while the transition (1, ) specifies that the reaction increases the
reaction count x by 1.
The argument shape is used to define the range of reaction counts that
the solver will consider when solving the CME. Since initial_copies = 20,
the range of reaction counts considered for shape = (initial_copies + 1, )
is 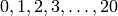 .
.
Finally, the argument initial_state defines
that the CME is initialised in the state (0, ). In other words,
at the initial time 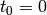 , the probability that there have been
zero occurrences of the reaction
, the probability that there have been
zero occurrences of the reaction 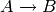 is 1.
is 1.
Note
You may be wondering why shape, initial_state, and the item contained by transitions have tuple values, not scalar values. This is because models typically represent systems containing more than one reaction.
Solving the model¶
We now create a solver s to solve the CME for the model m:
import numpy
from cmepy import solver
s = solver.create(
model = m,
sink = False
)
time_steps = numpy.linspace(0.0, 3.0, 6)
for t in time_steps:
s.step(t)
This code initialises the solver using solver.create and then advances the
solution of the chemical master equation from time 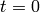 to time
to time
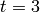 using 5 uniformly spaced steps.
using 5 uniformly spaced steps.
The attribute s.y stores the current solution of the CME computed by the solver. This solution is essentially a dictionary, mapping reaction counts to probabilities. For example, s.y[(3, )] is the probability that there have been exactly three occurances of the reaction.
Note
The keyword argument sink = False indicates that the state space of the model is not truncated, that is, there is no way probability may ‘leak’ out of the states under consideration of the solver.
Recording and plotting results¶
Now we add a recorder r to record the solutions and compute statistics:
from cmepy import recorder
r = recorder.create(
(('A->B', ), )
)
time_steps = numpy.linspace(0.0, 3.0, 6)
for t in time_steps:
s.step(t)
r.write(t, s.y)
The call to recorder.create initialises a recorder with a single random
variable named 'A->B'. By default, this random variable is the first
coordinate of the state space, which in this case is the reaction
count of the reaction 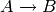 .
.
Solutions are written to the recorder by r.write(t, s.y), where t is the time of the solution, and s.y is the solution computed by the solver s.
The measurements recorded for the random variable named 'A->B' are stored inside r under the key 'A->B'. For example, the attribute r['A->B'].expected_value is a list of the expected values of the random variable 'A->B' that have been recorded by r.
We can now use the recorder r to generate the marginal distributions over the reaction counts, for each time step, and then plot these distributions using Matplotlib‘s pylab module:
import pylab
pylab.figure()
for t, d in zip(r['A->B'].times, r['A->B'].distributions):
marginal = d.to_dense(m.shape)
pylab.plot(marginal, label = 't = %.1f' % t)
pylab.xlabel('Reaction count')
pylab.ylabel('Probability')
pylab.legend()
pylab.savefig('simple_plot.png')
We access a measurement instance for the random variable 'A->B' via r['A->B']. This measurement instance contains the attributes times and distributions, which are the lists of measurement times and marginal distributions, respectively.
Each marginal distribution d is represented as a ‘sparse’ dictionary structure, which must be converted to a ‘dense’ flat array for plotting, using the method d.to_dense(shape). In this case, the shape of the array we want is the same shape of the model, m.shape.
Note
Python’s built-in zip(a, b) function takes two lists a and b, and returns the list [(a[0], b[0]), (a[1], b[1]), ...].
Note
You may like to experiment using pylab.show() instead of pylab.savefig('simple_plot.png').
Bringing everything together¶
Mixing all the above ingredients together yields the following Python script:
import numpy
import pylab
from cmepy import model, solver, recorder
def main():
initial_copies = 20
m = model.create(
propensities = [lambda x: initial_copies - x],
transitions = [(1, )],
shape = (initial_copies + 1, ),
initial_state = (0, )
)
s = solver.create(
model = m,
sink = False
)
r = recorder.create(
(('A->B', ), )
)
time_steps = numpy.linspace(0.0, 3.0, 6)
for t in time_steps:
s.step(t)
r.write(t, s.y)
pylab.figure()
for t, d in zip(r['A->B'].times, r['A->B'].distributions):
marginal = d.to_dense(m.shape)
pylab.plot(marginal, label = 't = %.1f' % t)
pylab.xlabel('Reaction count')
pylab.ylabel('Probability')
pylab.legend()
pylab.savefig('simple_plot.png')
if __name__ == '__main__':
main()
Suppose this script is saved as simple.py. Then we can run the script from the command line via:
python simple.py
This will save a plot of the reaction count distributions to simple_plot.py.
Note
If Python complains with the message ImportError: No module named cmepy, check that CmePy has been installed.